Wk03: Spreadsheet Best Practices & Data Import
Or, How I Learned to Stop Worrying and Think Like R
Learning objectives
- Understand how to format raw data to be “tidy”
- Know common data formatting/data entry mistakes to avoid
- Introduction to dates as data
- Know how to load a package
- Be able to describe what data frames and tibbles are
- Know how to use
read.csv()to read a CSV file into R - Be able to explore a data frame
- Be able to use subset a data frame
- Know the distinctions base R and
tidyverse
Part 1: Best Practices for Spreadsheets
This lesson is adapted from Data Carpentry’s Data Organization in Spreadsheets for Ecologists.
Formatting
- Never modify your raw data. Always make a copy before making any
changes.
- Keep track of all of the steps you take to clean your data in a plain text file.
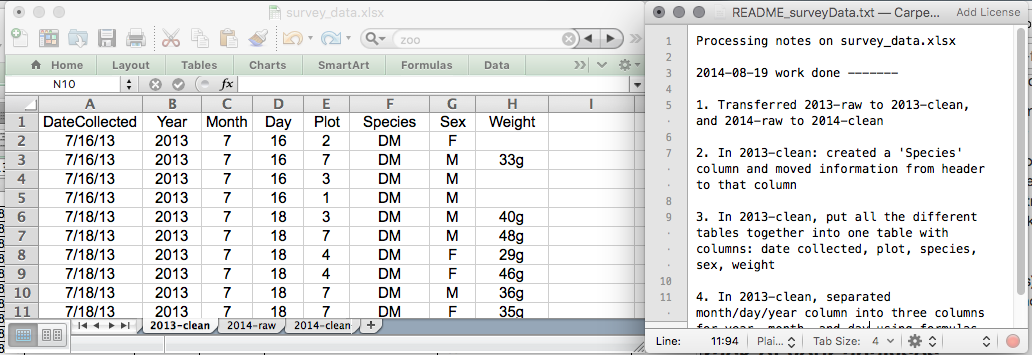
- Organize your data according to tidy data
principles.
- Each column is a variable like “weight” or “site_ID”
- Each row is a single observation
- Don’t combine pieces of information in a single cell
This is bad:
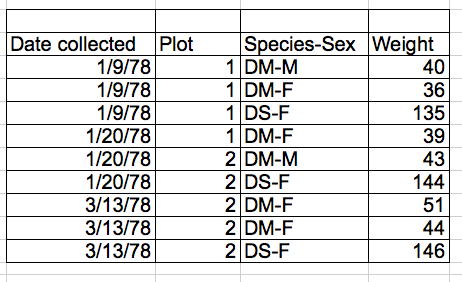
Why? Species and sex should be split into separate columns.
Avoiding Common Mistakes
Avoid using multiple tables within one spreadsheet.
Avoid spreading data across multiple tabs.
- Programs other than Excel will have a tough time seeing tabs
- Instead of adding a new tab, keep adding rows, and maybe use a new column for the info you’d be splitting across tabs. For example, use a column for Year instead of a new tab for a new year.
- If your table gets big, freeze your column headers to make them easier to see while you scroll.
Record zeros as zeros.
- Zeros are data! Don’t use blanks, and don’t use zeros as null values
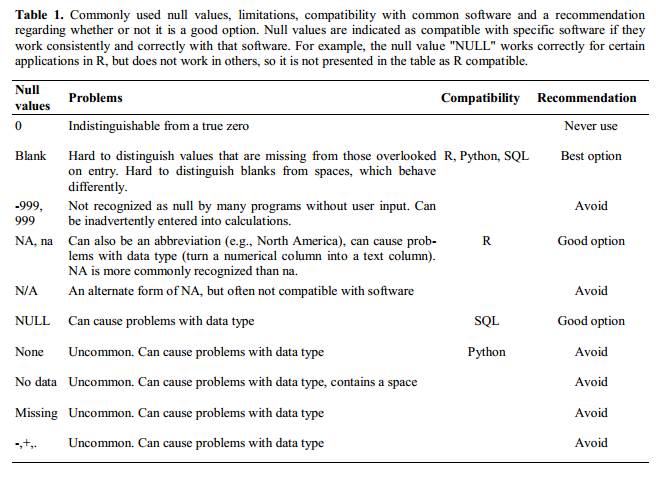
Use an appropriate null value to record missing data.
- There’s a broader discussion of this in White et al 2013, but here’s a quick look at their recommendations:
Don’t use formatting to convey information or to make your spreadsheet look pretty.
- R won’t see cell colors or bold fonts
- Keep data as characters or numbers
Place comments in a separate column.
- Don’t use Excel’s “comments” functionality, other programs don’t recognize these
- Just use a separate text column
Record units in column headers.
- Don’t put units in your data cells!
- 10cm will be read as characters, not numbers
Include only one piece of information in a cell.
Avoid spaces, numbers and special characters in column headers.
Good Name
Good Alternative
Avoid
Max_temp_C
MaxTemp
Maximum Temp (°C)
Precipitation_mm
Precipitation
precmm
Mean_year_growth
MeanYearGrowth
Mean growth/year
sex
sex
M/F
weight
weight
w.
cell_type
CellType
Cell Type
Observation_01
first_observation
1st Obs
- Avoid special characters in your data.
- Copy-pasting text into cells (ex. from Word) can do weird stuff
- Record metadata in a separate plain text file.
- When in doubt, use a README
- Put metadata in same directory as the data file itself, with an appropriate name
Dates as Data
- Excel was apparently created by people who hate time
- Excel stores dates in several different ways
- For Windows, it stores dates as number of days since Jan 1 1900
- For Mac, it stores dates as number of days since Jan 1
1904
- Check for dates that are ~4 years off
- Excel’s base is to store dates as numbers, but it dresses them up in different ways, like 9/1/2018 or Sept-1-2018
- Excel will often take one entry (Jul-10 for the 10th of July) and
interpret it as another (1-July-2010)
- It will even coerce data that aren’t dates into dates
- You have to be very careful, because Excel won’t tell you when it does things like this
- Treating dates as multiple pieces of data rather than one makes them
easier to handle.
- Use separate columns for year, month, and day
- You can also use columns for year and day-of-year (1 to 365)
- Ultimately, avoid letting Excel handle dates, it’s hard to know what it will do
Quality Control
- Always copy your original spreadsheet file and work with a copy so
you don’t affect the raw data.
- It can be helpful to have separate “Raw” and “Clean” data folders
- Use data validation to prevent accidentally entering invalid
data–this can be especially useful if you have others entering data for
you.
- Select the cells or column you want to validate
- On the
Datatab selectData Validation - In the
Allowbox select the kind of data that should be in the column. Options include whole numbers, decimals, lists of items, dates, and other values. - You can then give a range of values or enter a comma-separated list
of allowable values in the
Sourcebox - You can use the
Input Messagetab to customize the error message given when an incorrect value is entered
- “Sorry faithful undergraduate field assistant, we only have plots 1-10”
- You can make incorrect values throw warnings instead of errors using
the
Styleoption on theError Alerttab
- Use sorting to check for invalid data.
- Use conditional formatting (cautiously) to check for invalid data.
- Conditional formatting can color cells based on their value
- You can then visually scan for strange values
- Can be useful, but definitely not foolproof
Exporting Data
- Data stored in common spreadsheet formats will often not be read
correctly into data analysis software, introducing errors into your
data.
- try not to use .xls or .xlxs or .xxlsxlsxlllslxlsllaslxl or whatever
- Exporting data from spreadsheets to formats like CSV or TSV puts it
in a format that can be used consistently by most programs.
- Not only will this be easier for R to read, but it is widely recognized and will likely be around for a long time
- CSV and TSV are, ultimately, plain-text files, which are simple and classy and timeless
Part 2: Starting with Spreadsheets in R
Now that we’ve learned a bit about how R is thinking about data under the hood, using different types of vectors to build more complicated data structures, let’s actually look at some data.
Presentation of the Survey Data
We are studying the species repartition and weight of animals caught in plots in our study area. The dataset is stored as a comma separated value (CSV) file. Each row holds information for a single animal, and the columns represent:
| Column | Description |
|---|---|
| record_id | Unique id for the observation |
| month | month of observation |
| day | day of observation |
| year | year of observation |
| plot_id | ID of a particular plot |
| species_id | 2-letter code |
| sex | sex of animal (“M”, “F”) |
| hindfoot_length | length of the hindfoot in mm |
| weight | weight of the animal in grams |
| genus | genus of animal |
| species | species of animal |
| taxon | e.g. Rodent, Reptile, Bird, Rabbit |
| plot_type | type of plot |
Loading the Data
Your current R project should already have a data folder
with the surveys data CSV file in it. We can read it into R and assign
it to an object by using the read.csv() function. The first
argument to read.csv() is the path of the file you want to
read, in quotes. This path will be relative to your current
working directory, which in our case is the R Project
folder. So from there, we want to access the “data” folder, and then the
name of the CSV file. read.csv() also works with urls (if
you have an internet connection, of course). The url below links to a
csv stored on the course github page. You can call
read.csv() directly on the url:
surveys_url <- 'https://ucd-rdavis.github.io/R-DAVIS/data/portal_data_joined.csv'
surveys <- read.csv(surveys_url)or you can download the file and then load from your local computer.
Note that the destfile is a relative location, it assumes
there is a subdirectry called data in your working
directory, and then saves a file called
portal_data_joined.csv in that subdirectory.
download.file(url = surveys_url, destfile = 'data/portal_data_joined.csv')surveys <- read.csv('data/portal_data_joined.csv')- Hint: tab-completion works with file paths too. Type the pair of quotes, and with your cursor between them, hit tab to bring up the files in your working directory.
Take a look at your Environment pane and you should see an object called “surveys”. We can print out the object to take a look at it by just running the name of the object. We can also check to see what class it is.
surveys## record_id month day year plot_id species_id sex hindfoot_length weight
## 1 1 7 16 1977 2 NL M 32 NA
## 2 2 7 16 1977 3 NL M 33 NA
## 3 3 7 16 1977 2 DM F 37 NA
## genus species taxa plot_type
## 1 Neotoma albigula Rodent Control
## 2 Neotoma albigula Rodent Long-term Krat Exclosure
## 3 Dipodomys merriami Rodent Control
## [ reached 'max' / getOption("max.print") -- omitted 35546 rows ]class(surveys)## [1] "data.frame"Wow, printing a data frame gives us quite a bit of output. This is a lot more data than the small vectors we worked with last lesson, but the basic principles remain the same.
Data frames are really just a collection of vectors: every column is
a vector with a single data type, and every column is the exact same
length. You can make a data frame “by hand”, but they’re usually created
when you import some sort of tabular data into R using a function like
read.csv().
Inspecting data.frame Objects
When working with a large data frame, it’s usually impractical to try to look at it all at once, so we’ll need to arm ourselves with a series of tools for inspecting them. Here is a non-exhaustive list of some common functions to do this:
- Size:
nrow(surveys)- returns the number of rowsncol(surveys)- returns the number of columns
- Content:
head(surveys)- shows the first 6 rowstail(surveys)- shows the last 6 rowsView(surveys)- opens a new tab in RStudio that shows the entire data frame. Useful at times, but you shouldn’t become overly reliant on checking data frames by eye, it’s easy to make mistakes
- Names:
colnames(surveys)- returns the column namesrownames(surveys)- returns the row names
- Summary:
str(surveys)- structure of the object and information about the class, length and content of each columnsummary(surveys)- summary statistics for each column
Note: most of these functions are “generic”, they can be used on
other types of objects besides data.frame.
Challenge
Based on the output of str(surveys), can you answer the
following questions?
- What is the class of the object
surveys?
- How many rows and how many columns are in this object?
- How are our character data represented in this data frame?
- How many species have been recorded during these surveys? (This may
take more than just the
str()function. Try Googling around how to count the unique observations in a character string in R)
ANSWER
str(surveys)## 'data.frame': 35549 obs. of 13 variables:
## $ record_id : int 1 2 3 4 5 6 7 8 9 10 ...
## $ month : int 7 7 7 7 7 7 7 7 7 7 ...
## $ day : int 16 16 16 16 16 16 16 16 16 16 ...
## $ year : int 1977 1977 1977 1977 1977 1977 1977 1977 1977 1977 ...
## $ plot_id : int 2 3 2 7 3 1 2 1 1 6 ...
## $ species_id : chr "NL" "NL" "DM" "DM" ...
## $ sex : chr "M" "M" "F" "M" ...
## $ hindfoot_length: int 32 33 37 36 35 14 NA 37 34 20 ...
## $ weight : int NA NA NA NA NA NA NA NA NA NA ...
## $ genus : chr "Neotoma" "Neotoma" "Dipodomys" "Dipodomys" ...
## $ species : chr "albigula" "albigula" "merriami" "merriami" ...
## $ taxa : chr "Rodent" "Rodent" "Rodent" "Rodent" ...
## $ plot_type : chr "Control" "Long-term Krat Exclosure" "Control" "Rodent Exclosure" ...## * class: data frame
## * how many rows: 35549, how many columns: 13
## * the character data are characters if you have R Version 4.0.0 of later, factors for older versions
length(unique(surveys$species_id))## [1] 49table(surveys$species_id)##
## AB AH AS BA CB CM CQ CS CT CU CV DM
## 763 303 437 2 46 50 13 16 1 1 1 1 10596
## DO DS DX NL OL OT OX PB PC PE PF PG PH
## 3027 2504 40 1252 1006 2249 12 2891 39 1299 1597 8 32
## PI PL PM PP PU PX RF RM RO RX SA SC SF
## 9 36 899 3123 5 6 75 2609 8 2 75 1 43
## SH SO SS ST SU UL UP UR US ZL
## 147 43 248 1 5 4 8 10 4 2## * how many species: 48 (well, 49, but the 49th value is "", i.e., blank)Indexing and subsetting data frames
When we wanted to extract particular values from a vector, we used square brackets and put index values in them. Since data frames are made out of vectors, we can use the square brackets again, but with one change. Data frames are 2-dimensional, so we need to specify row and column indices. Row numbers come first, then a comma, then column numbers. Leaving the row number blank will return all rows, and the same thing applies to column numbers.
One thing to note is that the different ways you write out these indices can give you back either a data frame or a vector.
# first element in the first column of the data frame (as a vector)
surveys[1, 1] ## [1] 1# first element in the 6th column (as a vector)
surveys[1, 6] ## [1] "NL"# first column of the data frame (as a vector)
surveys[, 1] ## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25
## [26] 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50
## [ reached getOption("max.print") -- omitted 35499 entries ]# first column of the data frame (as a data.frame)
surveys[1] ## record_id
## 1 1
## 2 2
## 3 3
## 4 4
## 5 5
## 6 6
## 7 7
## 8 8
## 9 9
## 10 10
## 11 11
## 12 12
## 13 13
## 14 14
## 15 15
## 16 16
## 17 17
## 18 18
## 19 19
## 20 20
## 21 21
## 22 22
## 23 23
## 24 24
## 25 25
## 26 26
## 27 27
## 28 28
## 29 29
## 30 30
## 31 31
## 32 32
## 33 33
## 34 34
## 35 35
## 36 36
## 37 37
## 38 38
## 39 39
## 40 40
## 41 41
## 42 42
## 43 43
## 44 44
## 45 45
## 46 46
## 47 47
## 48 48
## 49 49
## 50 50
## [ reached 'max' / getOption("max.print") -- omitted 35499 rows ]# first three elements in the 7th column (as a vector)
surveys[1:3, 7] ## [1] "M" "M" "F"# the 3rd row of the data frame (as a data.frame)
surveys[3, ] ## record_id month day year plot_id species_id sex hindfoot_length weight
## 3 3 7 16 1977 2 DM F 37 NA
## genus species taxa plot_type
## 3 Dipodomys merriami Rodent Control# equivalent to head_surveys <- head(surveys)
head_surveys <- surveys[1:6, ] : is a special function that creates numeric vectors of
integers in increasing or decreasing order; try running
1:10 and 10:1 to check this out.
You can also exclude certain indices of a data frame using the
“-” sign:
surveys[, -1] # The whole data frame, except the first column## month day year plot_id species_id sex hindfoot_length weight genus
## 1 7 16 1977 2 NL M 32 NA Neotoma
## 2 7 16 1977 3 NL M 33 NA Neotoma
## 3 7 16 1977 2 DM F 37 NA Dipodomys
## 4 7 16 1977 7 DM M 36 NA Dipodomys
## species taxa plot_type
## 1 albigula Rodent Control
## 2 albigula Rodent Long-term Krat Exclosure
## 3 merriami Rodent Control
## 4 merriami Rodent Rodent Exclosure
## [ reached 'max' / getOption("max.print") -- omitted 35545 rows ]surveys[-c(7:34786), ] # Equivalent to head(surveys)## record_id month day year plot_id species_id sex hindfoot_length weight
## 1 1 7 16 1977 2 NL M 32 NA
## 2 2 7 16 1977 3 NL M 33 NA
## 3 3 7 16 1977 2 DM F 37 NA
## genus species taxa plot_type
## 1 Neotoma albigula Rodent Control
## 2 Neotoma albigula Rodent Long-term Krat Exclosure
## 3 Dipodomys merriami Rodent Control
## [ reached 'max' / getOption("max.print") -- omitted 766 rows ]Data frames can be subset by calling indices (as shown previously), but also by calling their column names directly:
surveys["species_id"] # Result is a data.frame
surveys[, "species_id"] # Result is a vector
surveys[["species_id"]] # Result is a vector
surveys$species_id # Result is a vectorIn general, when you’re working with data frames, you should make sure you know whether your code returns a data frame or a vector, as we see that different methods yield different results. Sometimes you get a data frame with one column, sometimes you get one vector.
You will probably end up using the $ subsetting quite a
bit. What’s nice about it is that it supports tab-completion! Type out
your data frame name, then a dollar sign, then hit tab to get a list of
the column names that you can scroll through.
Challenge
We are going to create a few new data frames using our subsetting skills.
- Create a new data frame called
surveys_200containing row 200 of thesurveysdataset. - Create a new data frame called
surveys_last, which extracts only the last row in ofsurveys.- Hint: Remember that
nrow()gives you the number of rows in a data frame - Compare your
surveys_lastdata frame with what you see as the last row usingtail()with thesurveysdata frame to make sure it’s meeting expectations.
- Hint: Remember that
- Use
nrow()to identify the row that is in the middle ofsurveys. Subset this row and store it in a new data frame calledsurveys_middle. - Reproduce the output of the
head()function by using the-notation (e.g. removal) and thenrow()function, keeping just the first through 6th rows of thesurveysdataset.
ANSWER
## 1.
surveys_200 <- surveys[200, ]
## 2.
# Saving `n_rows` to improve readability and reduce duplication
n_rows <- nrow(surveys)
surveys_last <- surveys[n_rows, ]
## 3.
surveys_middle <- surveys[n_rows / 2, ]
## 4.
surveys_head <- surveys[-(7:n_rows), ]Base R vs. tidyverse
Almost every time you work in R, you will be using different “packages” to work with data. A package is a collection of functions used for some common purpose; there are packages for manipulating data, plotting, interfacing with other programs, and much much more.
All of the stuff we’ve covered so far has been using R’s “base”
functionality, the built in functions and techniques that come with R by
default. There is a new-ish set of packages called the
tidyverse which does a lot of the same stuff as base R,
plus much much more. The tidyverse is what we will focus on
primarily from here on out, as it is a very powerful set of tools with a
philosophy that focuses on being readable and intuitive when working
with data. There are a few reasons we’ve taught you a bunch of base R
stuff so far:
- Base R can be quick and useful in a lot of ways
- The
tidyversestill works with the same building blocks as base R: vectors! - Some packages you need to use will only work with base R
- You will someday use Google and find a perfect solution to your problem, using base R
- You will probably have a collaborator at some point who uses base R
- The
tidyverseis constantly evolving, which can be good (new features!) and bad (really oldtidyversecode may behave differently when you update)
For example, using [] to subset data and using
read.csv() are base R ways of doing things, but we’ll show
you tidyverse ways of doing them as well.
In R, there are almost always several ways of accomplishing the same task. Showing you every single way of getting a job done seems like a waste of time, but we also don’t want you to feel lost when you come across some base R code, so that’s why there might be a bit of redundancy.
Loading Packages
Almost every time you work in R, you will be using different “packages” to work with data. A package is a collection of functions used for some common purpose; there are packages for manipulating data, plotting, interfacing with other programs, and much much more.
For much of this course, we’ll be working with a series of packages
collectively referred to as the tidyverse. They are
packages designed to help you work with data, from cleaning and
manipulation to plotting. They are all designed to work together nicely,
and share a lot of similar principles. They are increasingly popular,
have large user bases, and are generally very well-documented. You can
install the core set of tidyverse packages with the
install.packages() function:
install.packages("tidyverse")It is usually recommended that you do NOT write this code into a script, or the package will be reinstalled every time you run the script. Instead, just run it once in your console, and it will be permanently installed so you can use it any time.
Once a package has been installed on your computer, you can load it in order to use it:
library(tidyverse)Loading the tidyverse package actually loads a whole
bunch of commonly used tidyverse packages at once, which is pretty
convenient.
A common feature of tidyverse functions is that they use
underscores in the name. For example, the tidyverse
function for reading a CSV file is read_csv() instead of
read.csv(). Let’s try it:
t_surveys <- read_csv("data/portal_data_joined.csv")## Rows: 35549 Columns: 13
## ── Column specification ────────
## Delimiter: ","
## chr (6): species_id, sex, genus, species, taxa, plot_type
## dbl (7): record_id, month, day, year, plot_id, hindfoot_length, weight
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Now let’s take a look at how prints and check the class:
t_surveys## # A tibble: 35,549 × 13
## record_id month day year plot_id species_id sex hindfoot_length weight
## <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl>
## 1 1 7 16 1977 2 NL M 32 NA
## 2 2 7 16 1977 3 NL M 33 NA
## 3 3 7 16 1977 2 DM F 37 NA
## 4 4 7 16 1977 7 DM M 36 NA
## 5 5 7 16 1977 3 DM M 35 NA
## 6 6 7 16 1977 1 PF M 14 NA
## 7 7 7 16 1977 2 PE F NA NA
## 8 8 7 16 1977 1 DM M 37 NA
## 9 9 7 16 1977 1 DM F 34 NA
## 10 10 7 16 1977 6 PF F 20 NA
## # ℹ 35,539 more rows
## # ℹ 4 more variables: genus <chr>, species <chr>, taxa <chr>, plot_type <chr>class(t_surveys)## [1] "spec_tbl_df" "tbl_df" "tbl" "data.frame"Ooh, doesn’t that print out nicely? It only prints 10 rows by
default, NAs are now colored red, and under the name of each column is
the type of data! One important thing to notice is that the column types
are only double and character, no factors
here. By default, read_csv() keeps character data as
character columns, which would be like setting
stringsAsFactors=FALSE in read.csv().
Also, class() returned multiple things! You’ll notice
one of them is data.frame, but there are things like
tbl_df as well. The tidyverse has a special
type of data.frame called a “tibble”. Tibbles are the same
as data frames, but they print nicely as we just saw, and they usually
return a tibble when you’re using bracket subsetting. As always, just be
sure to check whether you’re getting a tibble or a vector back.
surveys[,1] # gives a vector back## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25
## [26] 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50
## [ reached getOption("max.print") -- omitted 35499 entries ]t_surveys[,1] # gives a tibble back## # A tibble: 35,549 × 1
## record_id
## <dbl>
## 1 1
## 2 2
## 3 3
## 4 4
## 5 5
## 6 6
## 7 7
## 8 8
## 9 9
## 10 10
## # ℹ 35,539 more rowsThis lesson is adapted from the Data Carpentry: R for Data Analysis and Visualization of Ecological Data Starting With Data materials.